Article Text
Abstract
Objectives Important findings can be masked in gene expression studies of mixed cell populations. We examined single-cell gene expression in SLE patient monocytes in the context of clinical and immunological features.
Methods Monocytes were purified from patients with SLE and controls, and individually isolated for single-cell gene expression measurement. A panel of monocyte-related transcripts were measured in individual classical (CL) and non-classical (NCL) monocytes.
Results Analyses of both CL and NCL monocytes demonstrated that many genes had a lower expression rate in SLE monocytes than in controls. Unsupervised hierarchical clustering of the CL and NCL data sets demonstrated independent clusters of cells from the patients with SLE that were related to disease activity, type I interferon (IFN) and medication use. Thus, each of these factors exerted a different impact on monocyte gene expression that could be identified separately, and a number of genes correlated uniquely with disease activity. We found within-cell correlations between genes directly induced by type I IFN-induced and other non–IFN-induced genes, suggesting the downstream biological effects of type I IFN in individual human SLE monocytes which differed between CLs and NCLs.
Conclusions In summary, single-cell gene expression in monocytes was associated with a wide range of clinical and biological features in SLE, providing much greater detail and insight into the cellular biology underlying the disease than previous mixed-cell population studies.
- Systemic Lupus Erythematosus
- Cytokines
- Autoimmunity
This is an Open Access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Statistics from Altmetric.com
Introduction
SLE is a severe chronic autoimmune disease that affects multiple organ systems, including the skin, joints and kidneys. Aberrant phenotype and function of monocytes is a prominent inflammatory feature associated with SLE.1–3 Classical (CL) monocytes are characterised by high expression of the CD14 but no significant CD16 expression (CD14++CD16−), and non-classical (NCL) monocytes are characterised by CD16 expression and intermediate to low CD14 expression (CD14+ CD16++).4 These subtypes have distinct biological activities,5 and NCL monocytes produce a greater amount of proinflammatory cytokines as compared with CL monocytes after stimulation.6 NCLs make up between 5% and 15% of the total monocyte population,7 and thus previous studies of all monocytes lumped together would likely miss any patterns specific to the NCL population.
A pivotal role for type I interferon (IFN) in the pathogenesis of SLE is strongly supported by many lines of evidence,8–10 and a type I IFN signature (overexpression of multiple type I IFN-induced transcripts) is a hallmark of gene expression studies of SLE in whole blood and mixed peripheral blood mononuclear cells (PBMCs).11–13 While this IFN signature has been observed in SLE for more than 15 years, progress in the cell biology of the IFN signature in the human immune system has been slow, as it is hard to interpret immune cell functions from the mixed-cell population data. Our group and others have examined gene expression in sorted cell populations in SLE, for example, studying monocytes, B cells and T-cell subpopulations.14 These studies have demonstrated that while each cell population may have an IFN signature, the transcripts which make up this signature differ between major immune cell populations.14 Thus far sorted, cell studies in SLE have examined major immune cell subsets, such as CD4 T cells, B cells, all monocytes, etc, and thus the mixed-cell population problem persists (eg, there are many functionally distinct T-cell subsets within the CD4 lineage, etc).
In mixed-cell population gene expression studies, when differences are observed it is not clear whether the observed differential gene expression was the result of differences that were uniformly present in all cells or whether they were the average of result of disparate differential expression results from various cell subsets.15 Cell type enumeration and deconvolution cannot fully address these issues, and thus, important biological subsets and within-cell correlations can be missed.16 In contrast, single-cell gene expression can reveal coupled transcriptional patterns within the same individual cell, which might allow for greater biological inference.17 In this study, we examined gene expression patterns in single CL and single NCL monocytes from patients with SLE. We observed clustering of specific cell populations within the CL and NCL monocyte subsets that corresponded to type I IFN, disease activity and medication usage. Each of these patterns was independent and distinct in our unbiased hierarchical clustering analysis, supporting the idea that each of these clinical factors induces a different change in monocyte subpopulations that can be detected using single-cell analysis. We also examined single-cell IFN signatures to determine what additional transcriptional events were present in the cells that were responding to type I IFN signalling. This allowed for an assessment of what happens to CL and NCL monocytes exposed to type I IFN in human patients with SLE in vivo, moving the IFN signature into cell biology in human disease.
Methods
Patients and samples
Peripheral blood samples from patients with SLE were obtained from the Mayo Clinic in Rochester, Minnesota, USA. Fifteen female patients were included in this study (see online supplementary table 1). All the cases fulfilled the American College of Rheumatology criteria for the diagnosis of SLE.18 Exclusion criteria included pregnancy, active chronic infection (eg, hepatitis C, HIV, etc), active acute infection, current intravenous therapy (eg, steroids or cyclophosphamide) or chemotherapy. Five healthy female controls matched for age were also studied. All the subjects provided informed consent, and the study was approved by the institutional review board.
Purification of classical (CD14++CD16−) and non-classical (CD14dimCD16+) monocytes
50 mL of peripheral blood was drawn from each subject into heparinised tubes. PBMCs were isolated over Ficoll-Paque Plus (GE Healthcare Bio-Sciences AB, Sweden). CD14++CD16− CL monocytes were purified by a magnetic separation, using the Human Pan-Monocyte Isolation Kit (Miltenyi) with modification of adding anti-CD16-biotin (Miltenyi) into the biotin–antibody cocktail, and a subsequent CD14+ selection (Miltenyi) was used to further increase purity. Purified CL monocytes were stained with Molecular Probes CellTracker Green CMFDA Dye (Life Technologies). CD14dimCD16+ NCL monocytes were purified similarly. CD16 microbeads (Miltenyi) were used during positive selection. Purity was checked by flow cytometry, and via this protocol, we achieved very high purity of both CL and NCL monocytes (see online supplementary figure 1). Cells were stained for CD56 and for human leukocyte antigen (HLA) DR. Natural killer (NK) cells were excluded by being HLADR− and CD56+ or dim.
C1 single-cell capture and pre-amplification
Single-cell capture and target gene pre-amplification were done using the Fluidigm C1 Single-Cell Auto Prep System and C1 Single-Cell Auto Prep Array Integrated Fluidic Circuits (IFCs), according to the manufacturer’s protocol. NCL and CL monocytes were loaded onto the C1 IFC sequentially, followed by visualisation using fluorescent microscope to determine NCL or CL lineage of each individual cell by direct visualisation (see online supplementary figure 2).
PCR gene expression in single cells
The target gene pre-amplified cDNAs were then assayed using 96.96 IFCs on the BioMark HD System from Fluidigm according to the manufacturer’s protocol. We designed primers for 87 of the target genes relevant to monocyte function, inflammation and cytokine signalling. Primers were all verified before being used in the single cell experiments. Primer sequences are included in online supplementary table 2. Melt curves were inspected individually to ensure that all PCR products were uniform. Individual amplification curves were also analysed, and those which did not follow the expected log-growth curve were also excluded. After this screening, duplicates were found to be within 5% of each other, similar to larger PCR reactions using a 384 well format.
Determination of genes directly induced by interferon-α in human monocytes, and interferon score calculation
Monocytes were resuspended at a final concentration of 2.5×106 cells per mL, cultured for 6 hours at 37°C with or without 100 U/mL IFN-α stimulation, and cDNA was made from mRNA. Real-time PCR was performed using the same 87 primers above and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an endogenous control. Gene expression was calculated as the difference cycle threshold (ΔCT) between the CT value of the target gene and the GAPDH. The genes with significant upregulation (p<0.05) after IFN-α stimulation were identified as genes directly induced by IFN-α (see online supplementary table 3). The sum of the expression values of these IFN-α-induced genes in each single monocyte was referred as the IFN score.
Statistics
Raw data were analysed using the Fluidigm Real-Time PCR Analysis software (V.4.1.2) with the linear (derivative) baseline correction method and a global CT threshold. All samples were normalised using the positive control mRNA spikes, as recommended.19–22 A failure score was calculated for each cell as outlined in ref 23. Cells with a failure score greater than mean+2xSD indicated that the overall expression level in that cell was very low, suggesting a dying cell, and these were excluded from analysis (see online supplementary figure 3). The limit of detection CT values was set at 28,24 and expression values were determined as 28—CT value. In this way, higher numbers correspond to greater gene expression, and CT values greater than or equal to 28 were considered non-detected. Overall gene expression for a given cell was calculated as the sum of expression values of all studied genes. Expression rate was calculated as the percentage of cells expresses a certain gene within a category (eg, SLE NCLs). Hierarchical clustering of gene expression values was done using JAVA Cluster (V.2.0) and visualised using Treeview software. Mann-Whitney U test was used to compare quantitative data between groups. In analyses that considered each of the 87 genes tested, we used a Bonferroni correction to establish a p value threshold for significance (p<5.75×10−4) to account for multiple comparisons.
Results
Gene expression rates in single monocytes of patients with SLE
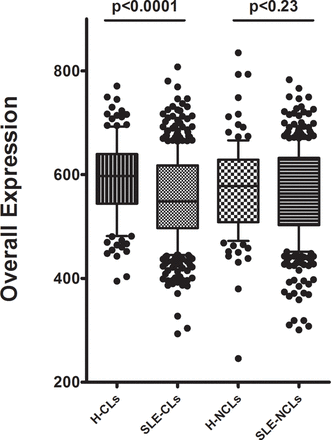
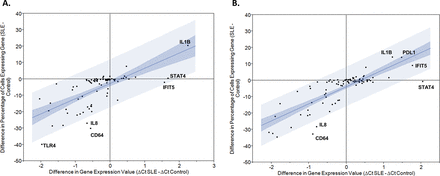
Gene expression was studied in 1113 individual monocytes (470 CL and 394 NCL from 15 patients with SLE and 130 CL and 119 NCL from 5 healthy donors). Individual monocytes showed a wide range of overall gene expression, measured as the sum of the expression values for each of the 87 transcripts (figure 1). Overall expression was higher in control CL monocytes as compared with SLE patient’s CL monocytes (p<0.0001). There was no significant difference in overall expression between NCL monocytes of patients with SLE compared with controls. Large differences in expression rates were observed between the various transcripts studied. For example, CD11c and transforming growth factor beta (TGFB) were expressed in less than 10% of the SLE CL monocytes, while Toll-like receptor (TLR8) and signal transducer and activator of transcription (STAT) 4 were expressed in more than 95% of the SLE CL monocytes (see online supplementary table 4). Comparing overall averaged quantitative expression between patients with SLE and controls, interferon-induced protein with tetratricopeptide repeats (IFIT) 5, STAT4 and interleukin (IL) 1B were significantly higher in SLE CLs, and programmed death-ligand (PDL) 1, IL-1B, IFIT5 and STAT4 showed significantly higher expression in SLE NCLs (figure 2). Comparing expression rates between patients with SLE and controls, IL-1B demonstrated significantly higher expression rate in CLs from patients with SLE versus healthy controls, and a similar non-significant trend was observed in NCLs (figure 2, see online supplementary table 4). Interestingly, many of the genes had either similar or lower expression rates in SLE monocytes as compared with healthy monocytes (figure 2). Those transcripts with significantly different expression rates between patients with SLE and controls in at least one of the two monocyte subtypes are shown in supplementary table 4. Expression rate correlated with the difference in overall averaged quantitative gene expression as would be expected. Interestingly, some transcripts varied from the 95% CI around the correlation line as shown in figure 2. These transcripts included IL-8 and CD64 in both CLs and NCLs, and TLR4 in CLs in which a much greater reduction in expression rate was observed compared with the decrease in overall averaged expression. This indicates that while fewer cells from patients with SLE expressed that transcript, the cells that did express the transcript in the patients with SLE did so at a higher quantitative level than the control cells. In NCL cells, STAT4 did not show any significant increase in expression rate but there was a significant increase in overall expression, again suggesting that the SLE cells that expressed this transcript did so at a higher level than the control cells that expressed the transcript. Notably, there were not any deviations above the 95% CI around the correlation line. This presents a new window into the gene expression in SLE immune cells, suggesting that cells which express particular inflammatory mediators may not be greater in number but instead have quantitatively greater expression in the same amount or even fewer number of cells expressing that transcript.
Overall gene expression in monocyte subsets in patients and controls. Overall expression is the sum of the CT values of all genes tested in that cell, boxes represent the IQR, line shows the median, error bars show the 10th and 90th percentile and p values are by Mann-Whitney U test. CLs, classical monocytes from healthy subjects; H, healthy subjects; NCLs, non-classical monocytes.
Plots showing the difference in expression rate versus the difference in averaged transcript amount between patients with SLE and healthy controls. Blue lines indicate the 95% CI around the correlation between the two parameters and notable transcript names are labelled on the graph. (A) Classical monocytes; (B) non-classical monocytes. CT, cycle threshold; CD64, cluster of differentiation 64; IFITS,interferon-induced protein with tetratricopeptide repeats 5,; IL1B,interleukin 1 beta,; IL8, interleukin 8,; PDL1, programmed death ligand 1; STAT4,signal transducer and activator of transcription 4 . Clustering analysis demonstrates subsets of Figure 1 monocytes corresponding to disease activity, interferon and medical therapy
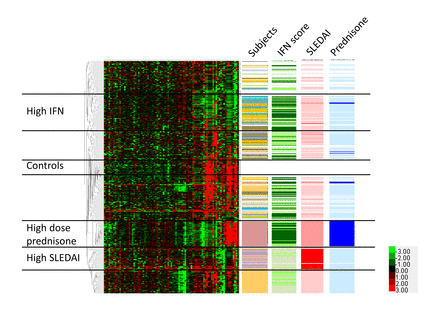
We next used unsupervised clustering analysis to examine patterns of correlation between cells and subgroups of cells within the CL and NCL populations. We used unsupervised clustering to provide an unbiased view of similarities in expression profile between cells. All NCLs from both patients with SLE and controls were analysed together, as shown in figure 3. Distinct clusters of cells could be observed within the NCL population, and these clusters were largely independent of the individual person from which the cells were derived. One exception is the individual who was on 20 mg of prednisone per day, forming a unique cluster of cells as indicated in figure 3. Interestingly, the majority of genes that clustered with the high-dose prednisone subject were downregulated, but both the PDL1 and TGFB genes were highly upregulated. Control cells were largely clustered in two areas, with one group in the central part of the clustering diagram that was separate from the patient cells, and a second group of control cells that comingled with the patient cells at the top of the diagram. Type I IFN as measured by the level of type I IFN-induced gene expression in the cell was also important to the overall clustering and, interestingly, IFN clusters were independent from the disease activity and prednisone-associated clusters. We also examined non-hierarchical clustering in the CL monocytes using the same strategy (see online supplementary figure 4). Patterns observed are similar, although notably the type I IFN cluster appears to be more diffuse. The patient receiving high-dose prednisone again clustered separately and, interestingly, the CL cells from that patient did not have as strong an IFN signature as the NCLs from the same patient. CL monocytes from patients with high systemic lupus erythematosus disease activity index (SLEDAI) scores were clustered together, similar to the NCL clustering diagram.
Hierarchical clustering of non-classical monocytes from patients and controls, with tracks indicating individuals, IFN score, SLEDAI score and prednisone usage. Each single cell forms a single row, and each column corresponds to an individual gene. All non-classical cells and genes are shown. Lower right corner inset shows heat map colour scheme key, with units indicating the delta CT values for each transcript. Colour codes for the data columns: Subject—white: healthy subjects; each colour represents one subject with SLE; interferon (IFN) score—darker green means higher IFN score; systemic lupus erythematosus disease activity index (SLEDAI) score—red: SLEDAI ≥10; pink: SLEDAI 3 to 6; white: SLEDAI 0 to 2; prednisone—dark blue: 20 mg/day; light blue: less than 10 mg/day; white: no prednisone. Black horizontal bars demarcate groups of cells that correspond to controls, high IFN, high SLEDAI and high-dose prednisone.
Differences in gene expression related to disease activity
We next examined which genes made up the set that differentiated the high disease activity of patients with SLE from low disease activity in CL and NCL monocytes. In CLs, janus kinase 1 (JAK1) and PDL1 showed a proportionate increase in both expression rate and overall expression in high SLEDAI subjects (figure 4A). In NCLs, PDL1 was significantly higher in both overall expression and expression rate, and chemokine (C-X-C motif) ligand (CXCL) 9 showed a decrease in expression rate with a concomitant increase in overall averaged expression (figure 4B). This suggests that quantitative CXCL9 expression was significantly higher in the NCLs expressing this transcript in the patients with high SLEDAI. As was observed in patients with SLE versus healthy controls comparison, there were many transcripts that showed a slight decrease in both averaged overall expression and expression rate. Online supplementary table 5 shows the genes with the most significant difference between low and high SLEDAI score patients in NCLs. Interestingly, TLR7 demonstrated an increase in expression rate but a decrease in overall averaged expression, suggesting that more cells expressed TLR7 in patients with high SLEDAI but at a lower level of expression. Transcripts showing the most dramatic decrease in expression rate in patients with high SLEDAI included STAT2, interferon alpha inducible protein (IFI) 27, CD32a, IL-8 and TLR4. Of these, only STAT2 showed a large (>6 CT) decrease in overall averaged quantitative expression. Thus, while the other transcripts may be expressed in fewer cells overall, those cells expressing these transcripts (IFI27, CD32a, IL-8, etc) did so at a high level, mitigating the effect of decreased expression rate on the overall averaged quantitative transcript level.
Plots showing the difference in expression rate versus the difference in averaged transcript amount between patients with SLE with SLEDAI ≥10 and those with SLEDAI 0–2. Blue lines indicate the 95% CI around the correlation between the two parameters, and notable transcript names are labelled on the graph. (A) Classical monocytes; (B) non-classical monocytes. CD32a = cluster of differentiation 32a, CD64 = cluster of differentiation 64, CXCL9 = chemokine (C-X-C motif) ligand 9, IFI27 = interferon alpha inducible protein 27, IFIT5 = interferon-induced protein with tetratricopeptide repeats 5, IL8 = interleukin 8, JAK1 = janus kinase1, PDL1 = programmed death ligand 1, SLEDAI = systemic lupus erythematosus disease activity index, STAT2 = signal transducer and activator of transcription 2, TLR4 = Toll-like receptor 4
Single-cell interferon signatures
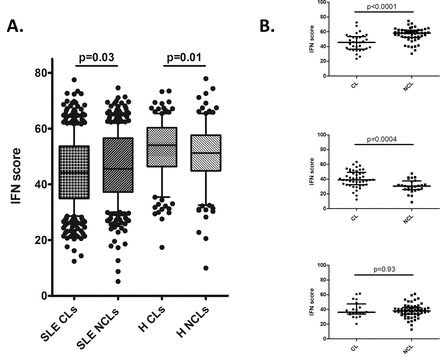
Using the single-cell data, we devised a method to examine the impact of type I IFN signalling in individual monocytes. We generated IFN scores based on the expression values of genes directly induced by type I IFN in human monocytes (see Methods). We used this score to estimate the relative recent impact of type I IFN on a given individual cell. Similar to the overall gene expression data, IFN score demonstrated a wide range of values in individual cells from the same blood sample (figure 5A). Interestingly, IFN scores were slightly higher in NCL monocytes than in CL monocytes in patients with SLE (figure 5A, p=0.03), and the opposite trend was observed in the healthy controls, in whom IFN scores were higher in CL cells. The healthy monocytes overall had higher IFN scores than the SLE patient monocytes, likely reflecting the overall decrease in expression we had observed for many transcripts (figure 2). Also, there were differences in CL versus NCL comparisons in individual patients as shown in figure 5B. The coexpression pattern of direct IFN-induced genes with other genes not directly induced by type I IFN offers insight into the effect of IFN on individual cells. We used non-parametric correlation analyses to analyse the relationship between IFN score and genes which are not directly induced by type I IFN. Interestingly, we observed different correlation patterns between these two monocyte subsets. The patterns were summarised into three models: transcripts that were correlated with IFN score in both CL and NCL monocytes, those that were correlated with IFN score in CL monocytes but not NCL monocytes and those that were correlated with IFN score in NCL monocytes but not CL monocytes (table 1). While the largest group of correlated transcripts is shared between both monocyte populations, there are also many transcripts that are correlated with IFN score in CL monocytes but not in NCL monocytes. Interestingly, these include interferon beta (IFNB) and a number of TLR transcripts. Two transcripts, 2'-5'-oligoadenylate synthase 1 (OAS1) and protein kinase R (PKR), are only correlated with IFN score in NCL monocytes but not in CL monocytes. These analyses document the diversity of IFN-related responses in the two monocyte populations, and emphasise the complex effects of IFN signalling in the various cells of the immune system.
Interferon (IFN) scores in monocyte subsets. (A) IFN score in all classical (CL) versus non-classical (NCL) monocytes from both patients with SLE and healthy controls. Boxes indicate the IQR, line shows the median and bars show the 10th and 90th percentiles. (B) Each graph shows the same comparison in an individual patient with SLE, each having a different pattern. Lines show the median and bars show the IQR. p Values by Mann-Whitney U test. H, healthy subjects.
Genes correlated with interferon (IFN) score in classicals (CLs) and non-classicals (NCLs)
Discussion
In this study, we demonstrate that patterns in single-cell gene expression in SLE monocytes reveal a wide range of biology, including the impact of type I IFN, medical treatment and autoimmune disease activity. There were some parallels but also many differences between the two closely related immune cell populations we studied—CL and NCL monocytes. This emphasises the complexity of the cellular immune system and suggests that much detail is lost when examining mixed-cell or bulk tissue gene expression data. When comparing patients and controls, we found greater differences in expression rate than in overall expression values for many genes, which suggests that these parameters are not completely correlated, and that biological effects could be mediated by changes in either parameter. These data would not be available in bulk-cell populations, and opens a new window into cellular biology. It was interesting that many genes were downregulated in patient samples as compared with controls, and this may reflect chronic activation of inflammatory pathways in the patients, which then causes downregulation of immune-related transcripts in peripheral circulating cells. Despite this, some transcripts were clearly upregulated in the patient samples, and these would be of particular interest in SLE monocytes.
A number of studies examining NCL function have been published.25 Several studies found that NCL monocytes have proinflammatory functions with higher expression of proinflammatory cytokines.26–28 However, CL monocytes also have inflammatory functions and have been shown to be the major producers of high levels of tumor necrosis factor alpha (TNF-α) and IL-1 in response to lipopolysaccharide (LPS).5 27 In NCL cells from the patient taking high-dose prednisone, expression of many inflammatory genes were decreased, including TNFA, IL-6, IL-8 and IL-1A and IL-1B. Meanwhile, the expression of TGFB and PDL1 were significantly increased. This is similar to other reports describing significantly elevated TGFB levels in monocytes from immunosuppressed patients,29 and these results could explain part of the immunosuppressive effect of prednisone in SLE.
Previous gene expression studies of bulk cells in patients with SLE have generally found a cross-sectional correlation between type I IFN-induced gene expression with disease activity,13 although this has not been robust in longitudinal studies.30 This suggests that type I IFN may indicate a patient group with a generally more severe disease, but that type I IFN may not fluctuate with disease activity over time. In our sample, type I IFN patterns were independent of both disease activity and medical treatment, supporting the idea that type I IFN is not a marker of disease activity in the moment. Previous studies of SLE monocytes examining correlations between gene expression and disease activity have suggested that both CD6431 and TLR432 are related to disease activity. In our study, we found that NCL cells from patients with high disease activity (SLEDAI ≥10) had a distinct pattern of gene expression, and these cells showed decreased expression of majority of genes but an increased expression of PDL1 and JAK1. It is possible that the upregulation of PDL1 in patients with SLE with high SLEDAI represents an immunoregulatory event in cells that have seen chronic immune stimulation. JAK1 being high in the CL monocytes from patients with high SLEDAI is also interesting, given the current trials of JAK inhibitors in patients with SLE. This study provides the first window into subsets of monocyte cells that are enriched in patients with SLE with high disease activity, and the results may assist in monitoring disease and advancing our understanding of the pathogenesis of SLE flare.
Acknowledgments
The authors would like to acknowledge the patients who contributed to this study, without their help this work could not have been done.
References
Footnotes
Contributors ZJ, WF, MAJ, JMD and GFB performed experiments, analysed data andcontributed to writing the manuscript. DMV, SA, AM, FE, TO, KM and VC recruited patients and collected patientdata, analysed data and contributed to the manuscript. TBN designed and supervised the study,revised the paper and performed data analysis and interpretation.
Funding WF: National Nature Science Foundation of China Grant No. 81402934; MAJ: Lupus Research Institute, Mayo Clinic Foundation; TBN: NIH grants (AR060861, AR057781, AR065964, AI071651), Rheumatology Research Foundation, the Mayo Clinic Foundation, the Lupus Research Institute, and the Lupus Foundation of Minnesota.
Competing interests None declared.
Ethics approval Mayo Clinic Institutional Review Board.
Provenance and peer review Not commissioned; externally peer reviewed.
Author note Zhongbo Jin and Wei Fan are co-first authors.
Correction notice The content of this article has been corrected since it first published Online First. Figure 3 has been replaced and the co-first authorship of the paper has been indicated.