Article Text
Abstract
Objective When faced with clinical symptoms of scarring alopecia—the standard diagnostic pathway involves a scalp biopsy which is an invasive and expensive procedure. This project aimed to assess if plucked hair follicles (HFs) containing living epithelial cells can offer a non-invasive approach to diagnosing inflammatory scalp lesions.
Methods Lesional and non-lesional HFs were extracted from the scalp of patients with chronic discoid lupus erythematosus (CDLE), psoriasis and healthy controls. RNA was isolated from plucked anagen HFs and microarray, as well as quantitative real-time PCR was performed.
Results Here, we report that gene expression analysis of only a small number of HF plucked from lesional areas of the scalp is sufficient to differentiate CDLE from psoriasis lesions or healthy HF. The expression profile from CDLE HFs coincides with published profiles of CDLE from skin biopsy. Genes that were highly expressed in lesional CDLE corresponded to well-known histopathological diagnostic features of CDLE and included those related to apoptotic cell death, the interferon signature, complement components and CD8+ T-cell immune responses.
Conclusions We therefore propose that information obtained from this non-invasive approach are sufficient to diagnose scalp lupus erythematosus. Once validated in routine clinical settings and compared with other scarring alopecias, this rapid and non-invasive approach will have great potential for paving the way for future diagnosis of inflammatory scalp lesions.
- cutaneous lupus
- scarring alopecia
- hair follicle
- diagnostic
- interferon-stimulated genes
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Introduction
Cutaneous lupus erythematosus (CLE) can present as part of the SLE spectrum or as a ‘skin only’ disease.1 Up to 80% of all CLE cases seen in dermatology settings are diagnosed to be chronic discoid lupus erythematosus (CDLE).2 In most patients with CDLE, lesions are predominantly in sun-exposed areas including face, neck, décolletage and, importantly, the scalp with often disfiguring lesions resulting in permanent scarring and irreversible hair loss.3–5 CLE in visible parts of the body is well recognised to significantly affect the quality of life of patients.6
Although CDLE can be diagnosed by experienced dermatologists based on morphological characteristics including erythema, pigmentary disturbances, telangiectasia and atrophy, a skin biopsy is usually required to confirm the diagnosis. Histopathological and immunohistological findings include interface dermatitis7 with a dense, predominantly CD8+ T-cell lymphocytic infiltrate particularly around the bulge region of the hair follicle (HF), basal cell/vacuolar degeneration representing apoptotic keratinocytes, basement membrane changes, infiltrating macrophages and plasmacytoid dendritic cells, pilosebaceous destruction and atrophic scarring.4 8–10 Many of these features are also seen in lichen planopilaris (LPP), a chronic inflammatory skin disease which also leads to permanent scarring alopecia.11–14 However, deposition of immunoglobulins and complement component 3 (C3) at the dermoepidermal junction as seen using direct immunofluorescence (DIF) is specific for CDLE but not LPP and thus a key differential diagnostic finding. Performing a scalp biopsy for histology and DIF is an invasive, thus often stressful procedure for the patient. It also requires availability of trained doctors and nurses, a special biopsy set-up with sterile equipment, patient’s consent regarding the side effects of local anaesthetic and invasive biopsy (including infection, scar, healing problems and bleeding) and usually a separate appointment, thus further time commitment for the patient. Furthermore, the biopsy needs to be processed, analysed and communicated by a dermatohistopathologist which takes up to 4 weeks in a standard dermatology setting. Thus, the overall costs involved to receive a conventional biopsy histology report are several hundred pounds/Euro (depending on the health system used).
A molecular hallmark of lupus erythematosus lesions is the high expression of interferon (IFN)-stimulated genes (ISGs).9 15 16 These include myxovirus protein A (MxA),17 IFN inducible protein 6 (IFI6), CXCL9, CXCL10, 2,5-oligoadenylate synthase (OAS) including OAS2 and OASL, IFN-induced helicase C domain-containing protein 1 (IFIH1/MDA5), bone marrow stromal antigen 2 (BST2) and guanylate-binding protein 1 (GBP1) in CLE lesions.8 18–21 It is well known that IFNs induce expression of major histocompatibility complex (MHC) class I and MHC class I-pathway related molecules (β2 microglobulin, β2M) and this has also been reported for organ-cultured human scalp skin.22–24 The proposed mechanism responsible for high expression of ISG includes the impaired clearance of apoptotic material, which results in secondary necrosis and release of immunostimulatory nucleic acids.25–27 An upregulated IFN response goes along with antiproliferative and antineovascularisation properties and may be involved in the downregulation of tissue repair mechanisms observed in CDLE.9 25 28 However, psoriasis which often presents with strong scalp involvement but never resulting in scars is also recognised to have some activation of the IFN pathway.29 In this work, we aimed to explore the potential of plucked HF analysis to identify CDLE-specific changes. We mainly focused on differentially expressed genes comparing HFs from lesional versus non-lesional areas of the scalp. Psoriasis samples were also included in our analysis, as a disease comparator which shows significant scalp inflammation with leukocyte infiltration but presents with different clinical outcomes regarding atrophy and scarring.30
Materials and methods
Patients
All patients had confirmed psoriasis as diagnosed by consultant dermatologists or CDLE (biopsy proven) and had been suffering long term (eg, longer than 5 years). All patients showed clinically active scalp lesions. Most patients with CDLE were on hydroxychloroquine but were asked to discontinue the drug for 3–5 days prior sampling. All patients with CDLE were female (age range 20–75 years of age, mean 55), 80% of healthy controls were female (mean age 46) and for patients with psoriasis, 50% were male and 50% female (mean age 43 years). Patients had not used topical corticosteroids within 48 hours prior to sampling. All individuals provided informed written consent and this research was carried out in compliance with the Declaration of Helsinki. The patients’ samples used for this study were collected under ethical approval, REC 10/H1306/88, National Research Ethics Committee Yorkshire and Humber–Leeds East. All experiments were performed in accordance with relevant guidelines and regulations.
Plucked HF samples
Four to five HFs from five healthy individuals (male and female aged 35–55 years), seven patients with CDLE from both lesional and non-lesional areas and six patients with psoriasis from both lesional and non-lesional areas were plucked out using tweezers as described previously.31 Only hairs with a full, visible HF were used (eg, telogen hairs or those with incompletely plucked follicle were excluded). Time to pluck four to five anagen hairs with suitable HFs usually takes an experienced person 5 min as up to 10 hairs have to be plucked to obtain this yield. In this study, we have restricted the area of ‘lesional’ sampling to scalp presenting with clinical signs of inflammation, such as erythema (including mild erythema), infiltration, hyperkeratosis/scaling. The hair shaft was cut-off and the white sheath (inner and outer root sheath) containing keratinocytes were used for this study (figure 3). Trimmed HFs were then immersed in Optimal Cutting Temperature (OCT) compound (Tissue-Tek; Sakura Finetek Europe, Alphen aan den Rijn, The Netherlands) in 1.5 mL tubes to preserve RNA quality before further processing. The samples were either stored in −80°C or processed immediately for RNA extraction.
Total RNA extraction from HFs
Plucked HFs were embedded longitudinally in OCT and 3–5 µm thick frozen sections were cut using a Leica CM3050S cryostat (Leica Microsystems, Buckinghamshire, UK) and collected in RNase free universal tubes. This was followed by the addition of lysis buffer (ReliaPrep RNA Cell Miniprep System; Promega, Wisconsin, USA) to the cut HF sections which were stored on dry ice prior to RNA extraction, which was carried out according to the manufacturer’s protocol. The quantity of the extracted RNA was measured using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, Massachusetts, USA), and the RNA quality was checked by an Agilent tape station Bioanalyser. RNA integrity number (RIN) number for RNA samples typically ranged between 7.5 and 10, with the majority of samples between 9 and 10, as determined at the Genomic Core Facility (GeneCore) in the European Molecular Biology Laboratory (EMBL, Heidelberg, Germany).
Transcriptomic and gene microarray analyses
At least 100 ng of the extracted total RNA was used as an input for microarray analysis done for each individual. Samples were processed at the EMBL GeneCore Facility for Affymetrix Microarray analysis (Geo number GSE119207). Gene Spring analysis was performed; in addition, Affymetrix CHP data were analysed with Transcriptome Analysis Console 3.1 software using HuGene-2_0-st-v1 library file from Affymetrix. A cut-off level of 1.5-fold change in the expression was used to analyse the differential gene expression when comparing lesional, non-lesional and healthy conditions. A selection of target genes identified by microarray was then validated via quantitative real-time PCR (qRT-PCR).
Quantitative real-time PCR
The mRNA expression of the target genes performed for each individual sample in this study was determined with qRT-PCR using a QuantStudio5 Real-Time PCR system (Applied Biosystems, Foster City, California, USA) using Qiagen QuantiTect SYBR Green Master Mix (Qiagen, Manchester, UK). 50–100 ng of total RNA was reverse transcribed using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Loughborough, UK) according to the manufacturer’s instructions. MxA, IFI6, IFIH1, CXCL9, CXCL10, BST2, OAS2, OASL, XAF1, BMP2, β2M, C3 and KDR QuantiTect primer assays were purchased from Qiagen (Hilden, Germany), whereas U6 snRNA32 primer (forward—5′-CTCGCTTCGGCAGCACA-3′ and reverse—5′-AACGCTTCACGAATTTGC-3′) was purchased from Sigma (Sigma-Aldrich, Poole, UK). The following parameters were used: initial heat activation, 95°C for 5 min; denaturation, 95°C for 10 s; combined annealing and elongation, 60°C for 30 s for a 40 cycle run. Data were analysed using the ΔΔCT method. mRNA expression of each gene of interest was normalised to U6snRNA housekeeping gene.
Statistical analysis of qRT-PCR results were analysed with GraphPad Prism software, V.7.00 (GraphPad Software, La Jolla, San Diego, California, USA). Results for each group are depicted as mean±SEM. Data were analysed by using one-way analysis of variance (ANOVA) followed by the Tukey multiple comparison test to determine statistically significant differences between groups: *p<0.05, **p<0.01.
Results
Microarray analysis of plucked HFs reveals a strong IFN signature and differential expression of complement, apoptosis and MHC I related genes in CDLE lesions
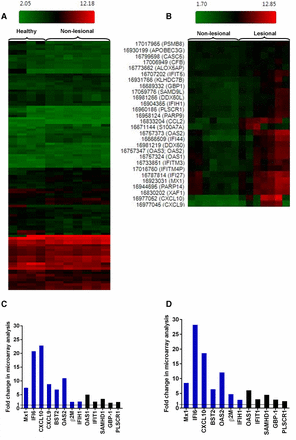
When comparing matched lesional and non-lesional CDLE samples for each patient as well as lesional CDLE with healthy samples, analysis of Affymetrix CHP data with Transcriptome Analysis Console showed substantial upregulation of ISGs, in particular Mx1, IFI6, BST2, OAS2 and CXCL10. This analysis also revealed significant upregulation between CDLE lesional samples compared with non-lesional or healthy samples of β2M, complement factor 3 (C3) and of genes involved in apoptotic cell death (XAF1, OAS2) which are also known to be IFN inducible (figure 1). Gene Spring analysis of the same microarray dataset indicated that the most robustly differentially expressed genes (p <0.005) in lesional versus non-lesional CDLE included β2M, TLR3, C1R, TAP1, SAMHD1, IRF1, XAF1, GBP1, OAS2, C3, IFI27, STAT1, CXCL10 and IFIH1/MDA5. Lesional CDLE samples showed the same set of genes consistently differentially expressed (p <0.005 for β2M, TLR3, C1R, TAP1, SAMHD1, XAF1, GBP1, OAS2, C3, IFI27, STAT1 and Mx1), when compared with healthy controls (online supplementary tables 1 and 2). Thus, with regard to differentially expressed genes associated with lupus pathology, such as ISGs, there is no significant overall difference between non-lesional and healthy samples as illustrated by the HeatMap overview (figure 1A) and figure 1B–D. However, subtle differences existed, such as lack of CXCL9 expression in healthy but not in non-lesional CDLE. Interestingly, β2M was significantly upregulated in non-lesional CDLE (p=3.80×10−4) compared with healthy samples in the Gene Spring analysis.
Supplemental material
Microarray analysis of plucked HF from healthy, lesional and non-lesional CDLE. (A) Hierarchical clustering comparing gene expression in HFs between healthy individuals and non-lesional CDLE and (B) between lesional and non-lesional CDLE. (C) Fold change of selected genes from microarray analysis comparing lesional with non-lesional CDLE, and (D) lesional CDLE with healthy. Genes which were also validated by qRT-PCR are indicated in blue. CXCL9 was not expressed in healthy samples and fold change can therefore not be given. CDLE, chronic discoid lupus erythematosus; HF, hair follicle; qRT-PCR, quantitative real-time PCR.
PCR analysis confirms that a set of genes related to IFN response, complement expression, apoptosis and MHC class I presentation characterises CDLE lesions
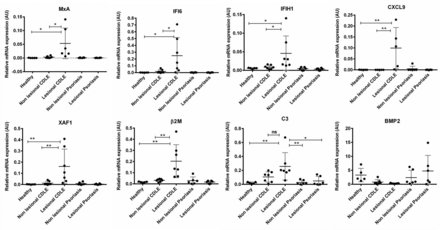
To confirm findings from the microarray analysis, qRT-PCR was performed on the same individual samples used in microarray analysis, as well as samples from patients with active scalp psoriasis. Expression of ISGs (Mx1, IFI6, CXCL9, CXCL10, IFIH1/MDA5), ISGs which are also apoptosis-related (XAF1), β2M and C3 were significantly higher in lesional CDLE as compared with lesional psoriasis or healthy samples (figure 2). Consistent with microarray data, no significant differences in ISGs expression between non-lesional CDLE and healthy samples were found. Interestingly, bone morphogenetic protein 2 (BMP2) was found downregulated in both lesional and non-lesional CDLE samples compared with psoriasis and/or healthy samples. Table 1 provides an overview regarding the fold changes of validated genes comparing CDLE with healthy samples and psoriasis with the same set of healthy samples. We failed to detect a marked IFN signature in psoriasis with the exception of slightly increased Mx1 and BST2. However, Kinase insert domain receptor (KDR) which codes for vascular endothelial growth factor receptor 2 (VEGFR2) was markedly upregulated in psoriatic samples but not significantly regulated in CDLE samples.
Differential gene expression in HF from lesional and non-lesional scalp. Selected genes were validated by qRT-PCR and expression compared between lesional and non-lesional psoriasis, CDLE as well as healthy individuals. Genes selected include IFN-stimulated genes (MxA, IFI6, CXCL9, IFIH1), apoptosis-related genes (XAF1), MHC class I antigen presentation pathway (β2M), complement factors (C3) and tissue repair-related gene (BMP2). Gene expression was normalised to housekeeping gene U6snRNA; arbitrary units are given. Except for BMP2, the same level of significance was also found between lesional CDLE and both psoriasis samples (not depicted for reason of clarity). β2M, β2 microglobulin; BMP2, bone morphogenetic protein 2; CDLE, chronic discoid lupus erythematosus; HF, hair follicle; IFI6, IFN inducible protein 6; IFN, interferon; MHC, major histocompatibility complex; MxA, myxovirus protein A; qRT-PCR, quantitative real-time PCR.
presents the fold change differences between genes validated by qRT-PCR when comparing average expression in lesional CDLE or psoriasis versus healthy control samples
Discussion
CDLE manifests with chronic inflammatory skin lesions which leads to permanent scarring, resulting in irreversible alopecia when affecting the scalp area.5 33 Early lesions can be confused with other inflammatory or infectious (eg, fungal) skin reactions, more advanced ones with scarring alopecias including primary scarring alopecias, such as LPP, alopecia mucinosa, folliculitis decalvans or secondary scarring alopecias such as morphea, cicatricial pemphigoid, neoplastic, traumatic or infection- associated granulomatous scarring alopecias. In clinical settings, the diagnosis can be difficult and histopathological confirmation is usually required.4 33 34 In this study, we performed gene expression analysis on plucked HFs from patients with CDLE and psoriasis, as well as healthy individuals. This pilot study showed that disease-specific signatures can be obtained from plucked HFs and this offers a promising, non-invasive, easy to perform approach with advantages over the use of skin biopsies. However, for diagnostic purposes it would be highly desirable to measure the identified signature molecules on the protein levels as this would allow for a higher sample stability, lower costs and less labour and time-consuming analysis. Our results revealed that ISG are significantly upregulated in lesional CDLE HFs and the pattern of differentially regulated genes resembles those found in previous reports on lesional full skin biopsies from patients with CDLE compared with healthy individuals or patients with psoriasis.8 35–37 Our finding of increased expression of the apoptosis-related genes, such as OAS2, OASL, XAF138–40 in lesional CDLE is also consistent with previously published reports using full skin biopsies.37 41 Our analysis demonstrates a distinct molecular signature for lesional CDLE with strong upregulation of ISG, apoptosis, complement and MHC I related genes which are hallmarks of the immunopathology in CDLE.1 8 20 42–44 CXCR3 ligands produced in response to epithelial IFNκ/IFNλ15 27 45 attract CD8+ cytotoxic T cells to the bulge area of the HFs leading to irreversible damage to the stem cell niche residing in this area, thus resulting in atrophic scarring.23 46–48 Within the normal bulge area, MHC class I and β2M are found downregulated and this stem cell niche is normally protected from inflammatory challenges, a phenomenon called ‘immune privilege (IP)’.22 23 49 In line with previous findings for scarring LPP,48 our data also point to IP collapse via increased expression of MHC class I (online supplementary table) and β2M in HF epithelium of lesional CDLE compared with non-lesional and healthy controls. Our microarray data show a slight tendency for downregulation of the key stem cell markers K15, CD200, Lhx2 and PHLDA1 residing within this area when comparing lesional CDLE with healthy/non-lesional CDLE (data not shown).
Regarding psoriasis, which can present with florid scalp inflammation, the HF IP is maintained and scarring hair loss is not seen although ISG have been found in psoriatic inflammation of the skin. This is in line with previous molecular analysis of scalp psoriasis highlighting it as an interfollicular disease.30 Supporting those findings, our analysis found low expression of inflammatory molecules including ISGs in HFs from patients with psoriasis. This suggests that the HF may be actively protected from inflammatory attack. In the psoriasis samples, KDR expression was higher compared with lesional CDLE (table 1). KDR is one of the two VEGFRs. Angiogenesis is due to the action of vascular endothelial growth factor, which is a key molecule in psoriatic skin.50 Reduced angiogenesis observed in CDLE may be due to high expression of GBP-1,20 which is well known for its action on new vessel formation. CDLE is characterised by atrophy and insufficient tissue repair response and the marked downregulation of BMP2, which was among the most downregulated genes found, is interesting in this context. BMPs are signalling molecules belonging to the transforming growth factor (TGFβ) superfamily.51 BMPs are implicated in a variety of pathophysiological processes in the skin including wound healing.51 52 Thus, increased cellular senescence, reduced angiogenesis due to high expression of GBP-1,20 as well as cytotoxic attack of the HF stem cell compartments and the impact of BMP2 on TGFβ pathways are likely to contribute to atrophic scarring seen in patients with CDLE.53 Once the HF is lost, consequences for tissue repair may deteriorate. Plikus et al reported that during the wound healing process, only those dermal cells which were adjacent to the regenerated HFs differentiated into lipid-laden newly formed adipocytes, but those in the hairless skin did not, suggesting that HFs are necessary to establish adipocyte precursors and normal wound repair.54 Similar to CDLE, LPP leads to permanent scarring14 and shares some molecular characteristics with CDLE,48 hence the differential diagnosis can be difficult.13 While LPP samples were not included in this pilot project, our qRT-PCR validation data showed a significant increase in C3 in lesional CDLE (figure 2), which is used together with immunoglobulin deposits at the basement membrane zone (dermoepidermal junction) to differentiate between patients with CDLE and LPP using DIF in skin biopsies.11–13 Keratinocytes have been shown to synthesise C3 in response to cytokines, such as CCL2 and IFNɣ.55 56 Interestingly, CCL2 was among the most upregulated genes in the microarray analysis (online supplementary data). Our results show only subtle differences between the non-lesional CDLE and healthy samples, suggesting non-lesional HFs of patients with CDLE are only mildly affected by the disease. Patients with CDLE recruited to this study followed measures of ultraviolet (UV) light protection. We have not yet analysed sun-exposed HFs from patients with LE, which could well show subclinical inflammation.
In summary, this pilot project shows great potential regarding the diagnostic value of analysed plucked HFs. Once validated and optimised for protein detection, this could offer a significant advantage in clinical settings where the diagnosis of inflammatory lesions in HF-rich areas, such as the scalp, is often difficult and delayed due to the need of diagnostic biopsies and histopathological assessment. With the results obtained so far, we propose a diagnostic panel (figure 3), using plucked HFs followed by analysis of selected molecules, which should allow distinguishing CDLE from other pathologies. However, this requires validation in a prospective clinical study and comparison with standard dermatohistology diagnosis. Using this approach has the potential to save cost and avoid invasive biopsies, but would also offer significant advantages for research into the pathophysiology of scarring alopecias by allowing repeated sampling in the context of environmental challenges (eg, UV) or therapeutic interventions.
Proposed pathogenically relevant genes overexpressed in lesional CDLE HF. Photographs show HF plucked from the scalp area and used for RNA extraction. A longitudinal HF section is shown in H&E stain highlighting outer root, inner root sheath and hair bulb. The green box shows differentially expressed genes in lesional CDLE HF samples versus non-lesional/healthy control samples. Dermatohistopathology features (right-hand pictures, CDLE biopsies from routine diagnosis, pictures provided by dermatohistopathology, Leeds) such as vacuolar degeneration (apoptosis–XAF1), lupus band (middle picture direct immunofluorescence; deposition of immunoglobulin and complement–C3), CD8+ T-cell immune responses (β2M, CXCL9), atrophy (BMP2) along with IFN-stimulated genes (MxA, IFI6, IFIH1) correspond to highlighted genes in lesional CDLE HFs. β2M, β2 microglobulin; BMP2, bone morphogenetic protein 2; CDLE, chronic discoid lupus erythematosus; HF, hair follicle; IFI6, IFN inducible protein 6; IFN, interferon; MxA, myxovirus protein A; qRT-PCR, quantitative real-time PCR.
References
Footnotes
Contributors Study design: MW, MJG, EMV; Clinical contribution: AB, MW, MJG, EMV, MYMY; Laboratory-based experiments: MSh, AAA, JP; Data analysis: MSh, MW, MSt, EMV; Contribution to critical discussion of results: MSt, NVB, MJG, SE, EMV, MW; Manuscript writing and revision: MSh, AAA, SE, MJG, MYMY, AB, NVB, MSt, EMV, MW.
Funding This project was mainly supported by a grant from Lupus UK with additional funding by Medical Research Council grant MR/M01942X/1. MYMY is funded as an National Institute for Health Research (NIHR) Doctoral Research Fellow and EMV is funded as an NIHR clinician scientist. This research is also supported by the NIHR Leeds Biomedical Research Centre.
Disclaimer The views expressed are those of the author(s) and not necessarily those of the National Health Service, the NIHR or the Department of Health.
Competing interests None declared.
Patient consent for publication Not required.
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement Data are available in a public, open access repository. All data relevant to the study are included in the article or uploaded as supplementary information.