Abstract
Depleting regulatory T cells (Treg cells) to counteract immunosuppressive features of the tumor microenvironment (TME) is an attractive strategy for cancer treatment; however, autoimmunity due to systemic impairment of their suppressive function limits its therapeutic potential. Elucidating approaches that specifically disrupt intratumoral Treg cells is direly needed for cancer immunotherapy. We found that CD36 was selectively upregulated in intrautumoral Treg cells as a central metabolic modulator. CD36 fine-tuned mitochondrial fitness via peroxisome proliferator-activated receptor-β signaling, programming Treg cells to adapt to a lactic acid-enriched TME. Genetic ablation of Cd36 in Treg cells suppressed tumor growth accompanied by a decrease in intratumoral Treg cells and enhancement of antitumor activity in tumor-infiltrating lymphocytes without disrupting immune homeostasis. Furthermore, CD36 targeting elicited additive antitumor responses with anti-programmed cell death protein 1 therapy. Our findings uncover the unexplored metabolic adaptation that orchestrates the survival and functions of intratumoral Treg cells, and the therapeutic potential of targeting this pathway for reprogramming the TME.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The RNA-Seq data for intratumoral Treg cells are available in the Gene Expression Omnibus database under accession code GSE139325. All relevant data are available from the corresponding author upon request.
References
Roychoudhuri, R., Eil, R. L. & Restifo, N. P. The interplay of effector and regulatory T cells in cancer. Curr. Opin. Immunol. 33, 101–111 (2015).
Delgoffe, G. M. et al. Stability and function of regulatory T cells is maintained by a neuropilin-1–semaphorin-4a axis. Nature 501, 252–256 (2013).
Saito, T. et al. Two FOXP3+CD4+ T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 22, 679–684 (2016).
Rech, A. J. et al. CD25 blockade depletes and selectively reprograms regulatory T cells in concert with immunotherapy in cancer patients. Sci. Transl. Med. 4, 134ra162 (2012).
Sutmuller, R. P. et al. Synergism of cytotoxic T lymphocyte-associated antigen 4 blockade and depletion of CD25+ regulatory T cells in antitumor therapy reveals alternative pathways for suppression of autoreactive cytotoxic T lymphocyte responses. J. Exp. Med. 194, 823–832 (2001).
Marabelle, A. et al. Depleting tumor-specific Treg cells at a single site eradicates disseminated tumors. J. Clin. Invest. 123, 2447–2463 (2013).
Teng, M. W. et al. Conditional regulatory T-cell depletion releases adaptive immunity preventing carcinogenesis and suppressing established tumor growth. Cancer Res. 70, 7800–7809 (2010).
Walter, S. et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat. Med. 18, 1254–1261 (2012).
Nishikawa, H. & Sakaguchi, S. Regulatory T cells in tumor immunity. Int. J. Cancer 127, 759–767 (2010).
Simpson, T. R. et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J. Exp. Med. 210, 1695–1710 (2013).
Curtin, J. F. et al. Treg depletion inhibits efficacy of cancer immunotherapy: implications for clinical trials. PLoS ONE 3, e1983 (2008).
Arce Vargas, F. et al. Fc effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell 33, 649–663.e4 (2018).
Miragaia, R. J. et al. Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity 50, 493–504.e7 (2019).
Arpaia, N. et al. A distinct function of regulatory T cells in tissue protection. Cell 162, 1078–1089 (2015).
Burzyn, D. et al. A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295 (2013).
Cipolletta, D. et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553 (2012).
Panduro, M., Benoist, C. & Mathis, D. Tissue Treg cells. Annu. Rev. Immunol. 34, 609–633 (2016).
Vignali, D. A., Collison, L. W. & Workman, C. J. How regulatory T cells work. Nat. Rev. Immunol. 8, 523–532 (2008).
Chaudhry, A. & Rudensky, A. Y. Control of inflammation by integration of environmental cues by regulatory T cells. J. Clin. Invest. 123, 939–944 (2013).
Campbell, D. J. & Koch, M. A. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol. 11, 119–130 (2011).
Zeng, H. et al. mTORC1 couples immune signals and metabolic programming to establish Treg-cell function. Nature 499, 485–490 (2013).
Yang, K. et al. Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature 548, 602–606 (2017).
Huynh, A. et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat. Immunol. 16, 188–196 (2015).
Gerriets, V. A. et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat. Immunol. 17, 1459–1466 (2016).
Weinberg, S. E. et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 565, 495–499 (2019).
Angelin, A. et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 25, 1282–1293 e1287 (2017).
Wang, H., Franco, F. & Ho, P. C. Metabolic regulation of Treg cells in cancer: opportunities for immunotherapy. Trends Cancer 3, 583–592 (2017).
Li, X. et al. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat. Rev. Clin. Oncol. 16, 425–441 (2019).
Plitas, G. et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity 45, 1122–1134 (2016).
Piconese, S. et al. Human OX40 tunes the function of regulatory T cells in tumor and nontumor areas of hepatitis C virus-infected liver tissue. Hepatology 60, 1494–1507 (2014).
Zappasodi, R. et al. Rational design of anti-GITR-based combination immunotherapy. Nat. Med. 25, 759–766 (2019).
He, N. et al. Metabolic control of regulatory T cell (Treg) survival and function by Lkb1. Proc. Natl Acad. Sci. USA 114, 12542–12547 (2017).
Beier, U. H. et al. Essential role of mitochondrial energy metabolism in Foxp3+ T-regulatory cell function and allograft survival. FASEB J. 29, 2315–2326 (2015).
Ho, P. C. et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015).
Siska, P. J. & Rathmell, J. C. T cell metabolic fitness in antitumor immunity. Trends Immunol. 36, 257–264 (2015).
Vannini, N. et al. The NAD-booster nicotinamide riboside potently stimulates hematopoiesis through increased mitochondrial clearance. Cell Stem Cell 24, 405–418 e407 (2019).
Ravnskjaer, K. et al. PPARδ is a fatty acid sensor that enhances mitochondrial oxidation in insulin-secreting cells and protects against fatty acid-induced dysfunction. J. Lipid Res. 51, 1370–1379 (2010).
Morino, K. et al. Regulation of mitochondrial biogenesis by lipoprotein lipase in muscle of insulin-resistant offspring of parents with type 2 diabetes. Diabetes 61, 877–887 (2012).
Fan, W. et al. PPARδ promotes running endurance by preserving glucose. Cell Metab. 25, 1186–1193.e4 (2017).
Pascual, G. et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45 (2017).
Ladanyi, A. et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 37, 2285–2301 (2018).
Miska, J. et al. HIF-1α is a metabolic switch between glycolytic-driven migration and oxidative phosphorylation-driven immunosuppression of Treg cells in glioblastoma. Cell Rep. 27, 226–237.e4 (2019).
Phan, A. T., Goldrath, A. W. & Glass, C. K. Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity 46, 714–729 (2017).
Liu, P. S. & Ho, P. C. Mitochondria: a master regulator in macrophage and T cell immunity. Mitochondrion 41, 45–50 (2018).
Pacella, I. et al. Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc. Natl Acad. Sci. USA 115, E6546–E6555 (2018).
Son, N. H. et al. Endothelial cell CD36 optimizes tissue fatty acid uptake. J. Clin. Invest. 128, 4329–4342 (2018).
Dammone, G. et al. PPARγ controls ectopic adipogenesis and cross-talks with myogenesis during skeletal muscle regeneration. Int. J. Mol. Sci. 19, E2044 (2018).
Meeth, K., Wang, J. X., Micevic, G., Damsky, W. & Bosenberg, M. W. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res. 29, 590–597 (2016).
Ho, P. C. et al. Immune-based antitumor effects of BRAF inhibitors rely on signaling by CD40L and IFNγ. Cancer Res. 74, 3205–3217 (2014).
Driscoll, W. S., Vaisar, T., Tang, J., Wilson, C. L. & Raines, E. W. Macrophage ADAM17 deficiency augments CD36-dependent apoptotic cell uptake and the linked anti-inflammatory phenotype. Circ. Res. 113, 52–61 (2013).
Cheng, W. C. et al. Uncoupling protein 2 reprograms the tumor microenvironment to support the anti-tumor immune cycle. Nat. Immunol. 20, 206–217 (2019).
Picelli, S. et al. Full-length RNA-Seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Nelson, J. W., Sklenar, J., Barnes, A. P. & Minnier, J. The START App: a web-based RNAseq analysis and visualization resource. Bioinformatics 33, 447–449 (2017).
Liu, P. S. et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994 (2017).
Acknowledgements
We thank L.-F. Lu and W.-L. Lo for critical reading and comments. We also thank Y. Maeda and H. Nishikawa for helpful discussion. P.-C.H. was supported in part by the SNSF (project grants 31003A_163204 and 31003A_182470), the Swiss Cancer Foundation (KFS-3949-08-2016), the Swiss Institute for Experimental Cancer Research (ISREC 26075483), a European Research Council Staring Grant (802773-MitoGuide), the Cancer Research Institute Clinic and Laboratory Integration Program award and the SITC-MRA Young Investigator Award. C.J. is supported by the SNSF (project grants PMPDP3_129022 and PZ00P3_161459). A.Z. is supported by the SNSF (project grant 320030_162575) and Cancer League Switzerland (KFS-3394-02-2014). R.S. and I.G. are supported by NIH funding (P01 HL46403, P01 HL087018 and R01 HL142152 to R.S. and HL45095 and HL073029 to I.G.). E.M. acknowledges funding from the Swiss Cancer Research Foundation (KFS-3681-08-2015-R). S.-M.F. acknowledges funding from an FWO grant and projects, as well as KU Leuven Methusalem co-funding. J.F.-G. is supported by an FWO postdoctoral fellowship. J.D.W and T.M. are supported by NIH funding (P30 CA008748 and R01 CA056821), Swim Across America, the Ludwig Institute for Cancer Research, the Parker Institute for Cancer Immunotherapy and the Breast Cancer Research Foundation. R.Z. is supported by the Parker Institute for Cancer Immunotherapy Bridge Scholar Award. We also appreciate the support provided by the Electron Microscopy Facility at the University of Lausanne and the Biomedical Sequencing Facility at the Research Center for Molecular Medicine of the Austrian Academy of Sciences.
Author information
Authors and Affiliations
Contributions
H.W. and P.-C.H. contributed to overall project design and wrote the manuscript. H.W., F.F., Y.-C.T., C.-H.T. and F.P. performed the in vitro and in vivo animal works and data analysis. X.X. and S.R.M. performed analysis of the RNA-Seq results. H.W., M.P.T., R.Z., J.D.W., T.M., C.J., I.S. and A.Z. conducted collection and flow cytometry analysis of the human samples. J.F.-G. and S.-M.F. supported the metabolomite analysis. R.S. and I.G. provided the hybridoma clone for anti-CD36 antibody production and CD36flox mice, respectively. E.M. provided samples of NSCLC murine models.
Corresponding author
Ethics declarations
Competing interests
H.W. and P.-C.H. are inventors on a patent application related to targeting CD36 in cancer immunotherapy. P.-C.H. is serving as a member of the scientific advisory board for Elixiron Immunotherapeutics and receiving research grants from Roche and Idorsia. J.D.W. is serving as a consultant for Adaptive Biotechnologies, Advaxis, Amgen, Apricity, Array BioPharma, Ascentage Pharma, Astellas, Bayer, BeiGene, Bristol-Myers Squibb. Celgene, Chugai, Elucida, Eli Lilly, F-Star, Genentech, Imvaq, Janssen, Kyowa Hakko Kirin, Kleo Pharmaceuticals, Linnaeus, MedImmune, Merck, Neon Therapeutics, Northern Biologics, Ono, Polaris Pharma, Polynoma, PsiOxus, PureTech, Recepta, Takara Bio, Trieza, Sellas Life Sciences, Serametrix, Surface Oncology, Syndax and Synthologic. J.D.W. received research support from Bristol-Myers Squibb, MedImmune, Merck and Genentech and has equity in Potenza Therapeutics, Tizona Pharmaceuticals, Adaptive Biotechnologies, Elucida, Imvaq, BeiGene, Trieza and Linnaeus. P.-C.H. received an honorarium from Pfizer and Chugai. J.D.W. received an honorarium from Esanex.
Additional information
Peer review information L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
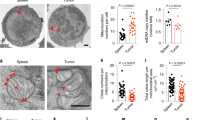
Extended Data Fig. 1 Lipid accumulation and increased CD36 expression in intratumoral Treg cells.
a, b, Quantitative results of geometric mean (GeoMean) fluorescent intensity of Bodipy FL C12 (a) and Bodipy 493/503 (Bodipy) (b) in Treg cells from paired TILs and PBMCs of non-small cell lung cancer (NSCLC) patients (n=6 per group). c, d, Quantitative results of GeoMean fluorescent intensity of Bodipy FL C12 (c) and Bodipy 493/503 (d) in Treg cells from paired PBMC and tumor infiltrated lymph nodes (TILNs) of melanoma patients (n=19). e, f, Quantitative results of fluorescent intensity of Bodipy FL C12 in Treg cells from indicated tissues of B16 melanoma-bearing mice (dLN, n=7; Spleen, n=6; Thymus, n=7; Tumor, n=6) (e), and MC38 colon carcinoma-bearing mice (n=8) (f). g, Quantitative result of GeoMean fluorescent intensity of CD36 surface staining in Treg cells from paired TILs and PBMCs of NSCLC patients (n=6) h, i, j, k, Quantitative results of surface expression of CD36 in Treg cells of indicated tissues from B16-OVA melanoma-bearing B6 mice (n=6, one outlier was removed from dLN) (h), inducible Braf/Pten melanoma-bearing mice (n=9) (i), K-rasLSL-G12D/+/p53fl/fl mouse model of NSCLC (Blood, n=8; Tumor, n=13) (j), and MC38 colon cancer (n=8) (k). l, CD36 expression in iTreg cells cultured in different indicated conditions for 48h. (RPMI: normal cell culture RPMI 1640 medium indicated in methods; CM, cancer cell conditioned medium, n=4 per group). m, CD36 expression in iTreg cells cultured in cancer cell-conditioned medium treated with control procedure or lipid removal procedure for 48h. (n=6 per group). Data are representative result of at least two independent experiments with similar results (l, m) or cumulative results from at least two independent experiments (a, b, c, d, e, f, g, h, i, j, k). Each symbol represents one individual. Data are mean ± S.D. and were analyzed by two-tailed, unpaired Student’s t-test (e, f, h, i, j, k, l, m) or two-tailed, paired Student’s t-test (a, b) or one-tailed, paired Student’s t-test (c, d, g).
Extended Data Fig. 2 CD36 expression supports the accumulation and suppressive function of intratumoral Treg cells.
a, Body weight of WT and TregCd36-/- mice at the age of 21–23 weeks (WT male, n=4; TregCd36-/- male, n=6; WT female, n=6; TregCd36-/- female, n=5;). b, Representative plots (left) and quantitative frequency of CD44hi/CD62Llow CD4+ or CD8+ T cells (right) in aged WT and TregCd36-/- mice (n=7 per group). c, Representative images of hematoxylin and eosin (H&E) staining for indicated tissues from WT or TregCd36-/- mice at the age of 21–23 weeks. Scale bar, 200 µm. d, e, Tumor growth of B16-OVA melanoma (n=7 per group) (d) or MC38 colon carcinoma (n=6 per group) (e) from WT or TregCd36-/- mice. f, g, h, Absolute number of FoxP3+ Treg cells per gram tumor (f), percentage of CD8+ T cells out of CD3+ T cells among tumor-infiltrating T cells (g), and the ratio of CD8+ to Treg cell TIL density (h) of YUMM1.7 melanoma-bearing WT and TregCd36-/- mice (n=11 per group). i, Representative plots (left) and percentage of indicated cytokine-producing CD4+ T cells among total tumor-infiltrating CD4+ T cells from indicated mice (right) (n=5 per group). Data are representative result of at least two independent experiments with similar results (c, d, e, i) or cumulative results from at least two independent experiments (a, b, f, g, h) Each symbol represents one individual. Data are mean ± S.D. (a, b, f, g, h, i) or ± S.E.M. (d, e) and were analyzed by two-tailed, unpaired Student’s t-test.
Extended Data Fig. 3 Effects of CD36 in expression of activation markers and stability of intratumoral Treg cells.
a, Representative images of guts (a) and spleens (b) from indicated group of Rag1-/- mice. c, d, e, Expression of CD44 (n=17) (c), CD103 (n=5) (d), and KLRG1 (n=5) (e) in intratumoral Treg cells of YUMM1.7 melanoma-bearing WT and TregCd36-/- mice. f, The expression of YFP in intratumoral Treg cells of YUMM1.7 melanoma-bearing WT and TregCd36-/- mice (n=15). g, h, i, Representative plots of IFNγ and TNF production among total intratumoral Treg cells from indicated mice (g), and quantitative result of percentage of IFNγ-producing (n=19 per group) (h) and TNF-producing (n=18, one outlier was removed from TregCd36-/-) (i) Treg cells among total intratumoral Treg cells of indicated mice. j, Expression of Ki67 in intratumoral Treg cells of YUMM1.7 melanoma-bearing WT and TregCd36-/- mice (n=10 per group). k, Representative histograms (left) and quantitative analysis (right) of Annexin V staining in intratumoral Treg cells from WT and TregCd36-/- tumor-bearing mice (n=14 per group). l, Quantitative analysis of cleaved caspase-3 levels in Treg cells of indicated tissues from WT (n=13 per group except for thymus, n=9) and TregCd36-/- (n=14 per group except for thymus, n=9) tumor-bearing mice. LN: non-draining lymph node; DLN: draining lymph node. Data are representative results of two independent experiments with similar results (a, b, d, e) or cumulative results from at least three independent experiments (c, f, g, h, i, j, k, l). Each symbol represents one individual. Data are mean ± S.D. (c, d, e, f, h, i, j, k, l) and were analyzed by two-tailed, unpaired Student’s t-test.
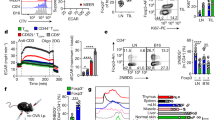
Extended Data Fig. 4 CD36-deficiency results in a metabolic shift and elevated apoptosis in Treg cells.
a, Indicated iTreg cells cultured in cancer cell-conditioned medium for 48 hrs (n=3 per group). Oxygen consumption rate (OCR) of indicated iTreg cells was measured and then followed by treatment with oligomycin, FCCP, and antimycin A plus Rotenone (n≥4 per group). b, Indicated iTreg cells cultured in cancer cell-conditioned medium for 48 hrs (n≥4 per group) and then media were refreshed with Seahorse Flux assay media without glucose. Basel extracellular acidification rate (ECAR) of indicated iTreg cells was measured and then followed by treatment with glucose, oligomycin, FCCP and 2-DG (n=4 per group). c, Quantitative result of glycolysis and glycolytic capacity based on the measurement of b. d, The viability of either WT or TregCd36-/- iTreg cells cultured under indicated conditions for 72 hrs (n=6 per group). Data are representative results of three independent experiments with similar results (a, b, c, d). Data are mean ± S.D. and were analyzed by two-tailed, unpaired Student’s t-test (c, d).
Extended Data Fig. 5 Intratumoral Treg cells require PPAR-β, not PPAR-γ, signaling for metabolic adaptation.
a, Enrichment plots of signals controlling mitochondrial matrix (left) and mitochondrial envelope in intratumoral Treg (n=4) compared to PBMC Treg cells (n=6), identified by GSEA computational method. ES: enrichment score; NES: normalized enrichment score; FDR: false discovery rate; NOM p-val: Nominal p value. b, c, d, Percentage of FoxP3+ Treg cells among CD4+ tumor-infiltrating T lymphocytes (n=5) (b), tumor growth (n=5) (c) and tumor weight (n=7) (d) from tumor-bearing WT and TregPPARγ-/- mice. e, Percentage of CD8+ T cells among tumor-infiltrating T cells from tumor-bearing WT and TregPPARβ-/- mice (n=10). f, Quantitative result of CD36+ intratumoral Treg cells from YUMM1.7 melanoma-bearing WT and TregPPARβ-/- mice (WT, n=14; TregPPARβ-/-, n=11). g, NAD/NADH ratio of indicated iTreg cells cultured in cancer cell-conditioned medium with DMSO or PPAR-β agonist for 48h (DMSO, n=8; PPAR-β agonist, n=10). Data are representative results of at least two independent experiments with similar results (b, c, d) or cumulative results from at least two independent experiments (e, f, g). Each symbol represents one individual. Data are mean ± S.D. (b, d, e, f, g) or ± S.E.M. (c) and were analyzed by two-tailed, unpaired Student’s t-test.
Extended Data Fig. 6 CD36-targeting unleashes host antitumor immunity.
a, b, c, d, Absolute number of FoxP3+ Treg cells per gram tumor (n=10 per group) (a), percentage of CD8+ T cells among tumor-infiltrating T cells (n=10 per group) (b) and representative plots and percentage of indicated cytokine-producing CD8+ T cells among total tumor-infiltrating CD8+ T cells (c) and CD4+ T cells among total tumor-infiltrating CD4+ T cells (d) from YUMM1.7 melanoma-bearing mice treated with indicated treatments (n=10 per group). e, f, Tumor growth (e) and survival curves (f) of YUMM1.7 melanoma-bearing B6 mice treated with indicated treatments (Ctrl, n = 10; α-PD1, n = 10; α-CD36, n = 11; α-CD36 + αPD-1, n = 11). Arrows indicate the date of treatment. Dotted lines indicate the tumor volume of 800 mm3. Data are cumulative results from at least two independent experiments. Each symbol represents one individual. Data are mean ± S.D. and were analyzed by two-tailed, unpaired Student’s t-test (a, b, c, d). Difference between survival curves was analyzed by Log-rank (Mantel-Cox) test (f).
Supplementary information
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Rights and permissions
About this article
Cite this article
Wang, H., Franco, F., Tsui, YC. et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat Immunol 21, 298–308 (2020). https://doi.org/10.1038/s41590-019-0589-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0589-5
This article is cited by
-
Modulating ferroptosis sensitivity: environmental and cellular targets within the tumor microenvironment
Journal of Experimental & Clinical Cancer Research (2024)
-
Targeting JMJD1C to selectively disrupt tumor Treg cell fitness enhances antitumor immunity
Nature Immunology (2024)
-
Immunosurveillance encounters cancer metabolism
EMBO Reports (2024)
-
Fatty acid metabolism of immune cells: a new target of tumour immunotherapy
Cell Death Discovery (2024)
-
Regulatory T cell-mediated immunosuppression orchestrated by cancer: towards an immuno-genomic paradigm for precision medicine
Nature Reviews Clinical Oncology (2024)